This article will describe the R-package “dataRetrieval” which simplifies the process of finding and retrieving water data from the U.S. Geological Survey and other agencies.
Package Overview
dataRetrieval is available on Comprehensive R Archive
Network (CRAN).
install.packages("dataRetrieval")Once the dataRetrieval package has been installed, it
needs to be loaded in order to use any of the functions:
library(dataRetrieval)There are several vignettes included within the
dataRetrieval package. The following command will open the
main package introduction:
vignette("dataRetrieval", package = "dataRetrieval")Additionally, each function has a help file. These can be accessed by typing a question mark, followed by the function name in the R console:
?read_waterdata_dailyEach function’s help file has working examples to demonstrate the usage. The examples may have comments “## Not run”. These examples CAN be run, they just are not run by the CRAN maintainers due to the external service calls.
Finally, if there are still questions that the vignette and help
files don’t answer, please post an issue on the
dataRetrieval GitHub page:
Orientation
dataRetrieval provides US water data mainly via 3
sources:
- USGS APIs (Water Data)
- Water Quality Portal (WQP)
- Water Services from National Water Information System (NWIS) - Legacy system that will eventually be retired.
Functions in dataRetrieval look like
read_waterdata_daily,
read_waterdata_continuous, readWQPqw,
summarize_waterdata_samples, etc. What does that mean? The
functions are generally structured with a prefix, middle, and
suffix:
-
Prefix: “read” or “what”
- “read” will access full data sets
- “summarize” will access data availability
-
Middle: “waterdata”, “WQP”, “NWIS:
- Water Data (waterdata) functions are the functions that get USGS water data. These pull from modern USGS API services.
- WQP functions are for discrete water-quality data from the Water Quality Portal.
- NWIS functions get data from legacy NWIS water services.
-
Suffix: “data” or other:
- Functions that end in “data”: These are flexible, powerful functions that allow complex user queries.
- Functions that don’t end with “data” are user-friendly functions that assume site, code, and start/end dates are known.
Data Overview
There are many types of data available from
dataRetrieval. To understand how the services are
separated, it’s helpful to understand that each data type is retrieved
from a completely different web service or API.
NWIS has traditionally been the source for all USGS water data
-
NWIS Water Services will be retired (scheduled late 2026, but uncertain):
-
USGS water data functions will slowly replace NWIS functions
-
read_waterdata_sampleshas replacedreadNWISqw -
read_waterdata_dailycan replacereadNWISdv -
read_waterdata_monitoring_locationcan replacereadNWISsite -
read_waterdata_ts_metacan replacewhatNWISdata -
read_waterdata_parameter_codescan replacereadNWISpCode -
read_waterdata_continuouscan replacereadNWISuv
-
-
Discrete water quality data:
- WQP functions should be used when accessing non-USGS discrete water quality data
-
read_waterdata_samplesshould be used for USGS data
USGS Water Data APIs
| data_type_cd | Legacy Function | Data description | New Function |
|---|---|---|---|
| uv | readNWISuv |
Continuous data | read_waterdata_continuous] |
| dv | readNWISdv |
Daily aggregated | read_waterdata_daily |
| gwlevels | readNWISgwl |
Groundwater levels | read_waterdata_field_measurements |
| site | readNWISsite |
Site metadata | read_waterdata_monitoring_location |
| pcode | readNWISpCode |
Parameter code metadata | read_waterdata_parameter_codes |
| stat | readNWISstat |
Site statistics |
read_waterdata_stats_por,
read_waterdata_stats_daterange
|
| rating | readNWISrating |
Rating curves | None yet |
| peak | readNWISpeak |
Peak flow | None yet |
| use | readNWISuse |
Water Use | None yet |
| meas | readNWISmeas |
Discrete surface water | read_waterdata_field_measurements |
readNWISdata |
General data import | read_waterdata |
Water Data API key
Do you regularly use dataRetrieval? As you switch your
workflows over to the new Water Data API functions, consider grabbing
yourself an API key, which will bump your limit up to 1,000 requests per
hour. Head to the sign
up page to get a token.
One you have your API key, add it to your .Renviron file
like this:
Replace [your api key] with the alphanumeric code provided by the
sign-up page. That’s it! dataRetrieval will look for an
.Renviron file in your directories and use it for making
web service calls.
USGS Basic Retrievals
The USGS uses various codes for basic retrievals. These codes can have leading zeros, therefore they need to be a character (“01234567”).
- Site ID (often 8 or 15-digits)
- Parameter Code (5 digits)
- Full list:
read_waterdata_parameter_codes()
- Full list:
- Statistic Code (for daily values)
- Full list:
read_waterdata_metadata("statistic-codes")
- Full list:
Here are some examples of a few common parameter codes:
|
|
Use the read_waterdata_parameter_codes function to get
information on USGS parameter codes.Then use your favorite data analysis
methods to pull out what you need. Here is one example to find all the
phosphorous parameter codes:
pcode <- read_waterdata_parameter_codes()
phosCds <- pcode[grep("phosphorus",
pcode$parameter_name,
ignore.case = TRUE
), ]Explore the wide variety of parameters that contain “phosphorus” in the parameter_name:
User-friendly retrievals: NWIS
Sometimes, you know exactly what you want. If you know:
- The type of data (groundwater, unit values, daily values, etc..)
- USGS site number(s)
- USGS parameter code(s)
- Time frame (start and end date)
You can use the “user-friendly” functions. These functions take the same 4 inputs (sites, parameter codes, start date, end date), and deliver data from the different NWIS services.
Pheasant Branch Creek Example
Let’s start by asking for discharge (parameter code = 00060) at a site right next to the old USGS office in Wisconsin (Pheasant Branch Creek).
siteNo <- "USGS-05427948"
pCode <- "00060"
start.date <- "2023-10-01"
end.date <- "2024-09-30"
pheasant <- read_waterdata_daily(monitoring_location_id = siteNo,
parameter_code = pCode,
time = c(start.date, end.date))From the Pheasant Creek example, let’s look at the data. The column names are:
names(pheasant)## [1] "monitoring_location_id" "parameter_code" "statistic_id"
## [4] "time" "value" "unit_of_measure"
## [7] "approval_status" "last_modified" "qualifier"
## [10] "geometry" "time_series_id"Let’s make a simple plot to see the data:
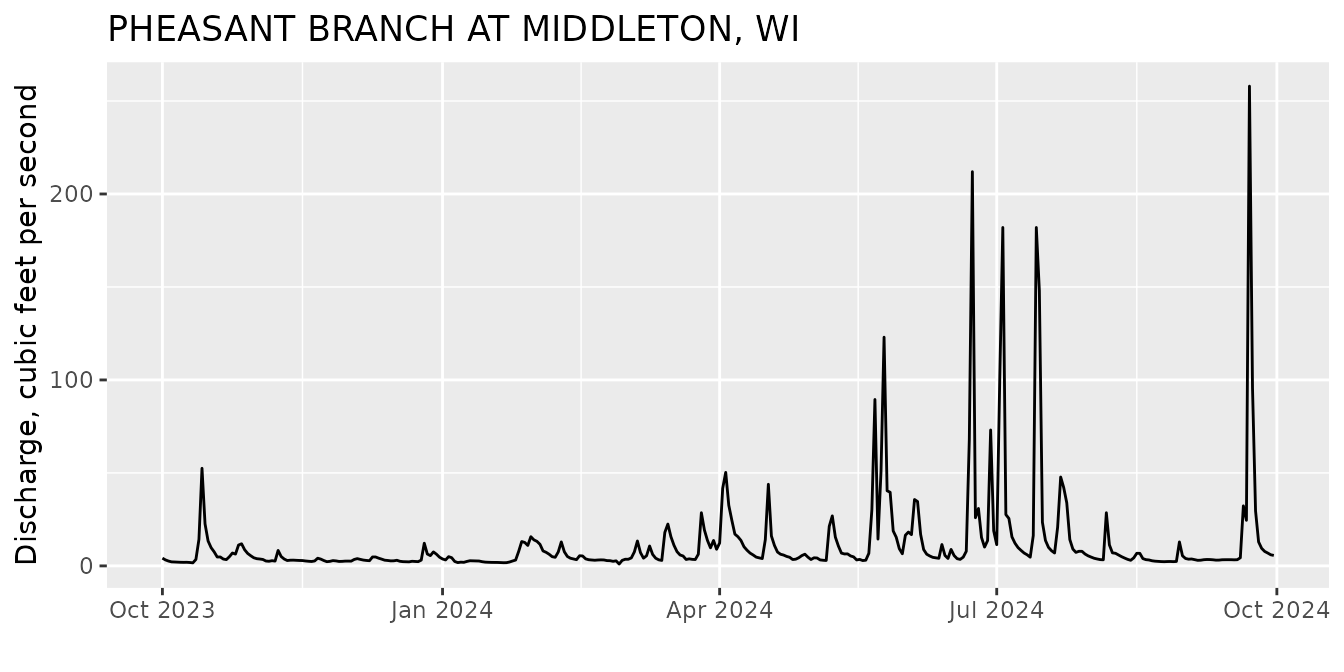
Then we can use the read_waterdata_parameter_codes and
read_waterdata_monitoring_location functions to create
better labels:
parameterInfo <- read_waterdata_parameter_codes(pCode)
siteInfo <- read_waterdata_monitoring_location(siteNo)
ts <- ts +
xlab("") +
ylab(parameterInfo$parameter_name) +
ggtitle(siteInfo$monitoring_location_name)
ts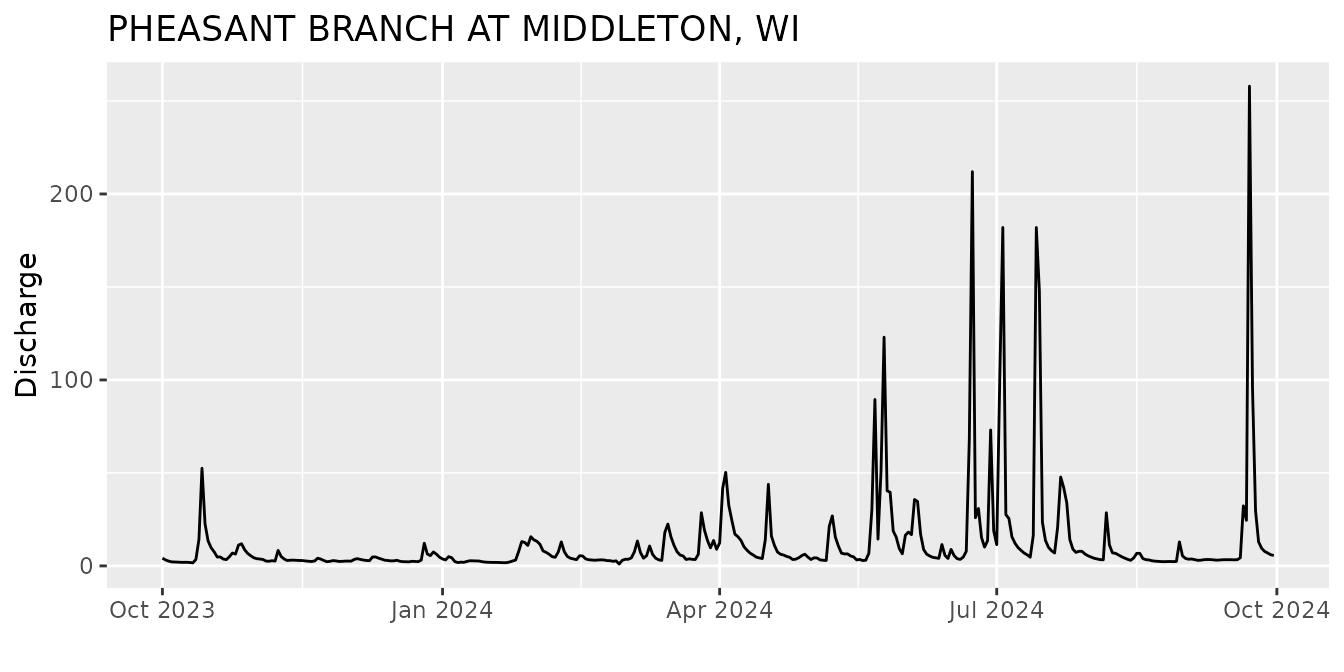
Known USGS site, unknown service/pcode
The most common question the dataRetrieval team gets is:
“I KNOW this site has data but it’s not coming out of dataRetrieval! Where’s my data?”
First verify that the data you think is available is actually
associated with the location. For time series data, use the
read_NWIS_ts_meta function to find out the available time
series data.
library(dplyr)
site <- "USGS-05407000"
ts_data_available <- read_waterdata_ts_meta(monitoring_location_id = site)
data_available <- ts_data_available |>
sf::st_drop_geometry() |>
mutate(begin = as.Date(begin),
end = as.Date(end)) |>
select(parameter_name, parameter_code, statistic_id, computation_identifier,
begin, end) The time series that have “Instantaneous” in the
computation_identifier column are available in the continuous data
service (read_waterdata_continuous), and the rest of the
data will be available in the daily service
(read_waterdata_daily).
dv_pcodes <- data_available$parameter_code[data_available$computation_identifier != "Instantaneous"]
stat_cds <- data_available$statistic_id[data_available$computation_identifier != "Instantaneous"]
stat_cds <- unique(stat_cds[!is.na(stat_cds)])
dv_data <- read_waterdata_daily(monitoring_location_id = site,
parameter_code = unique(dv_pcodes),
statistic_id = stat_cds)
uv_pcodes <- data_available$parameter_code[data_available$computation_identifier == "Instantaneous"]
uv_data <- read_waterdata_continuous(monitoring_location_id = site,
parameter_code = unique(uv_pcodes))
peak_data <- readNWISpeak(gsub("USGS-", "", site))For discrete water quality data, use the
summarize_waterdata_samples function:
discrete_data_available_all <- summarize_waterdata_samples(site)
discrete_data_available <- discrete_data_available_all |>
select(parameter_name = characteristicUserSupplied,
begin = firstActivity, end = mostRecentActivity,
count = resultCount)The discrete water quality data can be accessed with the
read_waterdata_samples function:
samples_data <- read_waterdata_samples(monitoringLocationIdentifier = site,
dataProfile = "basicphyschem")Water Quality Portal (WQP)
dataRetrieval also allows users to access data from the
Water Quality Portal. The
WQP houses data from multiple agencies; while USGS data comes from the
NWIS database, EPA data comes from the STORET database (this includes
many state, tribal, NGO, and academic groups). The WQP brings data from
all these organizations together and provides it in a single format that
has a more verbose output than NWIS.
This tutorial will use the modern WQX3 format. This is still considered “beta”, but it is the best way to get up-to-date multi-agency data.
The single user-friendly function is readWQPqw. This
function will take a site or vector of sites in the first argument
“siteNumbers”. USGS sites need to add “USGS-” before the site
number.
The 2nd argument “parameterCd”. Although it is called “parameterCd”, it can take EITHER a USGS 5-digit parameter code OR a characterisitc name (this is what non-USGS databases use). Leaving “parameterCd” as empty quotes will return all data for a site.
So we could get all the water quality data for site USGS-05407000 like this:
qw_data_all <- readWQPqw(siteNumbers = site,
parameterCd = "",
legacy = FALSE)or 1 parameter code:
qw_data_00095 <- readWQPqw(siteNumbers = site,
parameterCd = "00095",
legacy = FALSE)or 1 characteristic name:
qw_data_sp <- readWQPqw(siteNumbers = site,
parameterCd = "Specific conductance",
legacy = FALSE)Discover Data
This is all great when you know your site numbers. What do you do when you don’t?
There are 2 dataRetrieval functions that help with USGS
data discovery:
-
read_waterdata_monitoring_locationfinds sites within a specified filter -
read_waterdata_ts_metasummarizes the time series meta data
And 2 functions that help with discover in WQP:
-
readWQPsummarysummarizes the data available within the WQP by year. -
whatWQPdatasummarizes the data available within the WQP.
Available geographic filters are individual site(s), a single state,
a bounding box, or a HUC (hydrologic unit code). See examples for those
services by looking at the help page for the readNWISdata
and readWQPdata functions:
Here are a few examples:
Time/Time zone discussion
The arguments for all
dataRetrievalfunctions concerning dates (startDate, endDate) can be R Date objects, or character strings, as long as the string is in the form “YYYY-MM-DD”.-
For functions that include a date and time,
dataRetrievalwill take that information and create a column that is a POSIXct type. By default, this date/time POSIXct column is converted to “UTC”. In R, one vector (or column in a data frame) can only have ONE timezone attribute.- Sometimes in a single state, some sites will acknowledge daylight savings and some don’t
-
dataRetrievalqueries could easily span multiple timezones (or switching between daylight savings and regular time)
The user can specify a single timezone to override UTC. The allowable tz arguments are
OlsonNames(see also the help file forreadNWISuv).
Large Data Requests
It is increasingly common for R users to be interested in large-scale
dataRetrieval analysis. You can use a loop of either state
codes (stateCd$STATE) or HUCs to make large requests. BUT
without careful planning, those requests could be too large to complete.
Here are a few tips to make those queries manageable:
Please do NOT use multi-thread processes and simultaneously request hundreds or thousands of queries.
Take advantage of the
whatWQPdataandwhatNWISdatafunctions to filter out sites you don’t need before requesting the data. Use what you can from these faster requests to filter the full data request as much as possible.Think about using
tryCatch, saving the data after each iteration of the loop, and/or using a make-like data pipeline (for example, see thedrakepackage). This way if a single query fails, you do not need to start over.The WQP doesn’t always perform that well when there are a lot of filtering arguments in the request. Even though those filters would reduce the amount of data needed to transfer, that sometimes causes the pre-processing of the request to take so long that it times-out before returning any data. It’s a bit counterintuitive, but if you are having trouble getting your large requests to complete, remove arguments such as Sample Media, Site Type, these are things that can be filtered in a post-processing script. Another example: sometimes it is slower and error-prone requesting data year-by-year instead of requesting the entire period of record.
Pick a single state/HUC/bbox to practice your data retrievals before looping through larger sets, and optimize ahead of time as much as possible.
There are two examples scripting and pipeline that go into more detail.
But wait, there’s more!
There are two services that also have functions in
dataRetrieval, the National Groundwater Monitoring Network
(NGWMN) and Network Linked Data Index (NLDI). These functions are not as
mature as the WQP and NWIS functions. A future blog post will bring
together these functions.
National Groundwater Monitoring Network (NGWMN)
Similar to WQP, the NGWMN brings groundwater data from multiple
sources into a single location. There are currently a few
dataRetrieval functions included:
Network Linked Data Index (NLDI)
The NLDI provides a information backbone to navigate the NHDPlusV2 network and discover features indexed to the network. For an overview of the NLDI, see: https://rconnect.usgs.gov/dataRetrieval/articles/nldi.html
